Yesterday, I accomplished something that I believed was difficult, but after all wasn't: to develop the FDA and PMDA MedDRA validation rules in XQuery (it's easy if you know how).
The problem with MedDRA is that it is not open and public - you need a license. After you got one (I got a free one as I, as a professor in medical informatics, use MedDRA for research. Once you have the license, you can download the files. When I did, I expected some modern file format like XML or JSON or so, but to my surprise, the files come as oldfashioned ASCII (".asc") files with the "$" character as field separator. From the explanations that come with the files, it is said that the files can be used to build a relational database. However, the license does not allow me to redistribute the information in any form, so I could not build a RESFful web service that could then be used in the validation. As also the other validator just uses the ".asc" files "as is", I needed to find out how XQuery can read ASCII files that do not represent any XML.
I regret that MedDRA is not open and free for everyone (CDISC controlled terminology is). How can we ever empower patients to report adverse events when each patient separately needs to apply for a MedDRA license? This model is not of this time anymore...
The FDA and PMDA each contain about 20 rules that involve MedDRA. One of them is i.m.o. not implementable in software. Rule FDAC165/(PMDA)SD2006 states "MedDRA coding info should be populated using variables in the Events General Observation Class, but not in SUPPQUAL domains". How the hell can a software know whether a variable in a SUPPQUAL domain has to do with MedDRA? The only way I can see is that there is codelist attached to that variable pointing to MedDRA. If this is not the case, one can only guess (something computers are not so good in).
As MedDRA files are text files that do not represent XML, we cannot use the usual XQuery constructs to read them. Fortunately, XQuery 3.0 comes with the function "unparsed-text-lines()" which (among others) takes a file address as an argument. The file address however needs to be formatted as a URI, e.g.:
unparsed-text-lines('file:///e:/Validation_Rules_XQuery/meddra_17_1_english/MedAscii/pt.asc')
This function reads the file line by line. If it is then combined with the function "tokenize" which split strings in tokens based on a field separator, then XQuery can also easily read such oldfashioned text files. So the beginning of our XQuery file (here for rule FDAC350/SD2007), after all the namespace and functions declarations, looks like:
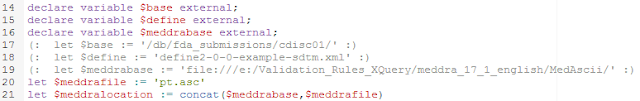
The first five lines in this part (18-22) define.the location of the define.xml file and of the MedDRA pt.asc (preferred terms file). For each of them, we use a "base" as we later want to enable that these are passed from an external program.
In line 24, the file is parsed, the result is an array of strings "$lines". In lines 26-29, we select the first item in each line (with the "$" character as the field separator). As such "$ptcodes" now simply consists of all the PT codes (preferred term codes).
Then, the define.xml file is read, and the AE, MH and CE dataset definitions are selected:
An iteration is started over the AE, MH and CE datasets (note that the selection allows for "splitted" datasets), and in each of them, the define.xml OID is captured of the --OID is captured, together with the variable name (which can be "AEPTCD", "MHPTCD" or "CEPTCD"). The location of the dataset is then obtained from the "def:leaf" element.
In the next part, we iterate over each record in the dataset, and get the value of the --PTCD variable (line 50):
and then check whether the value captured is one in the list of allowed PT codes (line 53). If it is not, an error message is generated (lines 53-55).
That's it! Once you know how it works, it is so easy: it took me about less than 15 minutes to develop each of these 20 rules.
I talked about these XQuery rules implementations with an FDA representative at the European CDISC Interchange in Vienna. When he saw the demo, his face became slightly pale, and he asked me: "Do you know what we paid these guys to implement our rules in software, and you tell me your implementation comes for free and is fully transparent?".
Beyond free, open and fully transparent (if that were not sufficient) the advantage of these rules is that the rules are completely independent of the software to do the validation: anyone can now write his own software without needing to code the rules themselves. You could even create a server that validates your complete repository of submissions during night time. As the messages come as XML, you can easily reuse them in any application that you want (try this with Excel!).
In the next section, I would like to explain how extremely easy it is to write software for executing the validations. The "Smart Dataset-XML Viewer" allows you to do these validations (but you can choose not do do any validation at all, or only for some rules), so I just took a few code snippets to explain this. We use the well-known open source Saxon library for XML parsing and validation, developed by the XML-guru Michael Kay, which is both available for Java and for C# (.NET). If you would like to see the complete implementation of our code, just go to the SourceForge site, where you can download the complete source code of just browse through. The most interesting class is the class "XQueryValidation" in the package "edu.fhjoanneum.ehealth.smartdatasetxmlviewer.xqueryvalidation".
Here is a snippet:
First of all, the file location of the define.xml is transformed to a "URI". A new StringBuffer is prepared to keep all the messages. In the following lines, the Saxon XQuery engine is initialized
and the base location and file name of the define.xml file is passed to the XQuery engine (remark that the define.xml can also be located in a native XML database, with one collection per submission, something that also the FDA and PMDA could easily do). This "passing" is done in the lines with "exp.bindObject" (in the center of the snippet).
In case MedDRA is involved in the rule execution, the same is done in the last part of the snippet (whether a rule required MedDRA is given by the "requiresMedDRA" attribute in the XML file containing all the rules:
The rule is then executed, and the error messages (as XML strings) captured in the earlier defined StringBuffer:
So, the contents of the "messageBuffer" StringBuffer is essentially an XML that can be parsed, or just written to file, or transformed to a table, or stored in a native XML database, or ...
In order to accept the passing of parameters from the program to the XQuery rule, we only need to change the hardcoded file paths and locations to "external" ones, i.e. stating that some program will be responsible for passing the information. In the XQuery itself, this is done by:
As one sees, lines 17-19 have been commented out, and lines 14-16 are lines 14-16 declare that the values for the location of the define.xml file and of the directory with MedDRA files will come from a calling program.
In the "Smart Dataset-XML Viewer", the user can himself decide where the MedDRA files are located (so it is not necessary to copy files to the directory of the application), using the button "MedDRA Files Directory":
A directory chooser than shows up, allowing to set where the MedDRA files need to be read from. This can also be a network drive, as is pretty usual in companies.
If you are interested in implementing these MedDRA validation rules, just download them from our website, or use the RESTful web service to get the latest update.
Again, the "Smart Dataset-XML Viewer" is completely "open source". Please feel free to use the source code, to extend it, to use parts of it in your own applications, to redistribute it with your own applications, etc.. Of course, we highly welcome it when you also donate source code of extensions that you wrote back, so that we can further develop this software.